Tutorials¶
Tutorial 1 - Introduction¶
File creation and basic write access¶
To create an empty file, we simply construct a new file handle, specifing the file path and a creation mode. For this example, we choose the truncate mode, which ensures, that the content of any pre-existing file is discarded. The code
file hdf5_file("first_file.h5", file::create_mode::truncate);
creates the empty HDF5 file ‘first_file.h5’ and names the resulting handle hdf5_file. With the first step done, we can add new objects to the file and store our data.
Before, we will store any data in our newly created file, we discuss one of the main benefits and major concepts of HDF5, the group. An HDF5 group allows us to save and access our data in a structured way. You can think of a group as an object similar to a folder in your favorite file system, since both are used to bundle related objects together, even other groups/folders. If you are familiar with UNIX file systems, you may even notice more similarities. Any new file starts with a group, which we call the root group and denote with ‘/’. Since the file and the root group are strongly related, we normally use the two terms interchangeable. This is reflected by the file class by providing the same interface as a group.
Using
group data = hdf5_file.create_group("data");
we create a new group ‘data’ within the root group and save the group handle in data.
Now, we will store some test data in our file. To do this, we have to create a new dataset. A dataset is a rectangular, homogeneous collection of elements and is the primary facility for data storage in HDF5. A dataset can distribute its elements over an arbitrary number of dimensions. This is convenient, since many objects in science, for example matrices and tensors, extend along several dimensions and we can map our data structures naturally onto our file structure.
dataset ds = data.create_dataset<double>("ds", { 10 });
will create a new dataset, named ‘ds’, of ten double elements along a single dimension and stores the dataset handle in ds. The final step is to store our data in the dataset, which can be done by using the, hopefully reasonable, syntax
ds <<= v;
In our example, v is a std::vector<double>, which contains ten elements. It should be noted, that the shape and datatype of the data must always match the shape and datatype of the dataset. An exception is thrown otherwise. While we will restrict ourself to the element type double and the container std::vector in this example, echelon supports several other types and containers, and can be extended to handle many more. Since this topic is a very complex one, we will discuss it in its entire at a later point. For now, it should be sufficient, that you know, that every primitive C++ type is supported.
The following code summarizes every step we have done so far, from the creation of our file to the storage of our data:
#include <echelon/echelon.hpp>
#include <vector>
using namespace echelon;
int main()
{
file hdf5_file("first_file.h5", file::create_mode::truncate);
group data = hdf5_file.create_group("data");
dataset ds = data.create_dataset<double>("ds", { 10 });
std::vector<double> v = {0, 1, 2, 3, 4, 5, 6, 7, 8, 9};
ds <<= v;
}
Basic read access¶
By now, we have learned how to store data in our file. But it would be rather pointless to store data in the file, if we could not load it again. In this section, we will learn how to load the data from the previous example into a container.
As a first step we create a handle
file hdf5_file("first_file.h5", file::open_mode::read_only);
to our old file ‘first_file.h5’. The only difference to the first example is, that we specify an open mode and not a creation mode. Since we only want to read data in this example, we use the read-only mode.
You may remember, that we stored our dataset within a group during the previous example. To access the dataset, we have to acquire a handle to the group first. For this purpose group overloads the square bracket operator, which takes the name of the object and returns a handle, if the object exists. Otherwise an exception is thrown. In our case we use
group data = hdf5_file["data"];
to acquire a group handle to the previously created group ‘data’.
The semantic of the above statement is much more complicated then one might expect at first glance. Since groups can store objects of many different types and the structure of the file is not available at compile-time, the square bracket operator does not know what kind of handle it should return. Instead it returns an object handle, which can reference any object. Therefore we call such a handle a polymorphic handle. To convert it to a meaningful handle, we guess a handle type at compile-time, in our case group, and assign the object handle to it. echelon then checks at runtime, if our guess does match the real type of the object and throws an exception, if we guessed wrong. One consequence of this is, that you can’t use the auto keyword in this case, since the object handle would not be converted to any meaningful type.
It is now straightforward to acquire a handle to the dataset
dataset ds = data["ds"];
using the same syntax and to load the data into the std::vector<double> container v using
auto_reshape(v) <<= ds;
While the container is automatically resized to hold the data if necessary, the value type of the dataset and the value type of the container must match again. Otherwise an exception is thrown.
Note
In general, echelon will not resize your containers, since they might not support this operation. To enable this functionality, you have to wrap your container in a call to auto_reshape as shown above.
The following code summarizes the second example and prints the content of the container using the standard output stream:
#include <echelon/echelon.hpp>
#include <vector>
#include <iostream>
using namespace echelon;
int main()
{
file hdf5_file("first_file.h5", file::open_mode::read_only);
group data = hdf5_file["data"];
dataset ds = data["ds"];
std::vector<double> v;
auto_reshape(v) <<= ds;
for(auto value : v)
std::cout << value << " ";
}
Tutorial 2 - Partial I/O and compression¶
Slicing¶
Until now, we have read entire datasets from the file into the main memory. While this is sufficient for simple use cases it is often not possible or desirable to read all data at the same time. For similar reasons, one might to assemble a dataset chunk by chunk instead of writing it in one go. In these use cases, partial I/O comes into play. It allows one to only read or write a certain subset of the dataset. Just now, echelon supports the use of regular, rectangular access patterns called slices. While slice usually only denotes a subset with a lower rank, we will also use it for selections with the same rank as the dataset for convenience. Creating slices in echelon is fairly trivial. For example the following code
using echelon::_;
auto slice = my_dataset(10, _);
will create a 1-slice (a one-dimensional slice) from a two-dimensional dataset by fixing the first index to ten. The slicing operator always takes the same number of arguments as the rank of the dataset. A runtime error is raised otherwise. echelon::_ is a placeholder for unrestricted dimensions. In a similar way, one can restrict a dimension to certain bounds. The code
using echelon::_;
using echelon::range;
auto slice = my_dataset(range(2, 10), _);
restricts the first dimension of the dataset to the interval 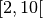 . If a certain bound should not be restricted the placeholder _ can be used.
For detailed informations about all possible slicing pattern, one should consult the documentation of the slicing operator.
. If a certain bound should not be restricted the placeholder _ can be used.
For detailed informations about all possible slicing pattern, one should consult the documentation of the slicing operator.
At the time of writing, echelon::slice does support most operations, which are valid on a dataset. For example, one can write data into the slice:
slice <<= data;
In addition, echelon supports slicing of array via the make_slice function. For example, the code
auto s = make_slice(c, _, range(1, 4));
slices the container c.
Array slices can be used to either write or read data from a dataset.
Warning
Currently, array slicing is only supported for mutable arrays and might not work otherwise.
As a convenience, echelon’s pre-defined containers support the same slicing syntax as dataset.
auto s = c(_, range(1, 4));
is equivalent to the previous example.
Compression¶
If working with large amounts of data it is often desirable to avoid a huge memory footprint, especially in the case of long-term storage. In many cases, one can achieve this by compressing the dataset, while sacrificing I/O performance at the same time. Luckily, echelon—respectively HDF5—comes with built-in compression capabilities. Compression can be enabled during dataset creation through the echelon::dataset_options class and the corresponding argument of the echelon::group::create_dataset() and echelon::group::require_dataset() methods by setting the respective option. For example
echelon::dataset_options options;
options.compression_level(4);
my_group.create_dataset<double>("test", {1000, 1000}, options);
will enable deflate compression with a compression level of 4.
HDF5 requires to change the storage layout of the dataset from ‘continuous’ to ‘chunked’ if compression is enabled. echelon will automatically try to guess a reasonable shape of the chunks. Since this choice might have a huge impact on compression ratio and I/O performance, one can override the auto-chunking heuristic by providing one’s own chunk shape through the echelon::dataset_options class. As of now, a single chunk should not be larger than the entire dataset.
Note
Enabling the shuffle filter via the corresponding option might increase the compression ratio.
Tutorial x - Advanced tidbits¶
Dimensions and dimension scales¶
One feature of datasets which wasn’t presented so far are dataset dimensions. The dimensions are accessed through the, hopefully indisputably named, dataset property dimensions. For example, one could give each dimension a descriptive label, as shown in the following code sample
my_dataset.dimensions()[0].relabel("foo");
For details, please refer to the corresponding API documentation.
Another useful feature linked to dataset dimensions are dimension scales which can be used to add certain meta-data to that dimension. For example, if one has saved a tabulated function in a dataset, one could add the corresponding value for each variable in a dimension scale, keeping this meta-data close to the actual data.
The following code shows how to add a dimension scale ‘x’ of type double to the first dimension of an already existing dataset
my_dataset.dimensions()[0].attach_dimension_scale<double>("x");
Echelon will automatically assemble the necessary data structures within the file and will free them if the dataset is destroyed. The shape of each dimensions scale is always one-dimensional and matches the length of the corresponding dataset dimension.
Since dimension scales are in a sense specialized datasets one can use them as is usual for datasets. For example the code
my_dimension_scale <<= x;
would write the content of the container x into the dimension scale.